Purity Independent Subtyping of Tumors (PurIST) delivers the first clinically robust, single-sample classifier for pancreatic ductal adenocarcinoma. Developed with UNC Lineberger collaborators, the classifier is resilient to tumor purity, batch effects, and assay platforms, enabling prospective use in routine pathology workflows.
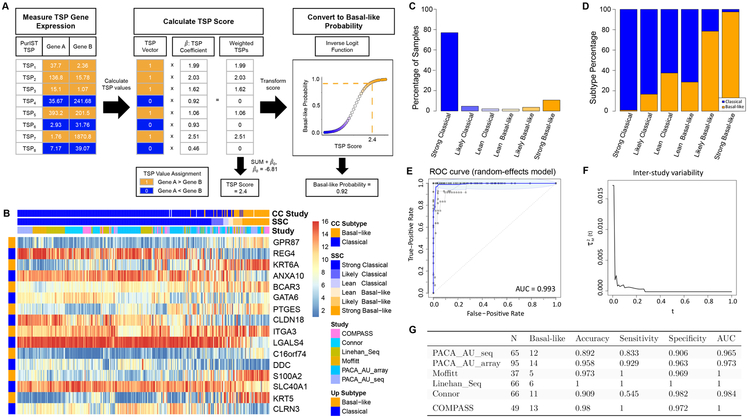
Development and validation of the PurIST SSC classifier across multiple independent cohorts, demonstrating robust performance regardless of tumor purity or sequencing platform.
From algorithm to diagnostic
- Methodological foundation. Rank-based scoring stabilizes gene-expression signatures in low-tumor-content samples, providing reproducible assignments across sequencing technologies.
- Rigorous validation. PurIST has been replicated across international cohorts, published in Clinical Cancer Research, and implemented in CLIA-certified laboratories supporting cooperative group trials.
- Clinical deployment. The test informs treatment selection in ongoing PDAC studies and is being evaluated as a stratification and enrichment tool for precision therapeutic trials.
Software and data resources
- Open-source training and inference code with example datasets are available to academic collaborators.
- Trial sponsors can access QA documentation and integration guidance via the Lineberger Biostatistics Shared Resource.
Key reference: (missing reference)