Projects
- Pancreatic cancer subtyping
- Statistical tools for high-throughput epigenomics
- Multi-study replicability of prediction models
- Unsupervised clustering of RNA-seq data
Pancreatic cancer subtyping
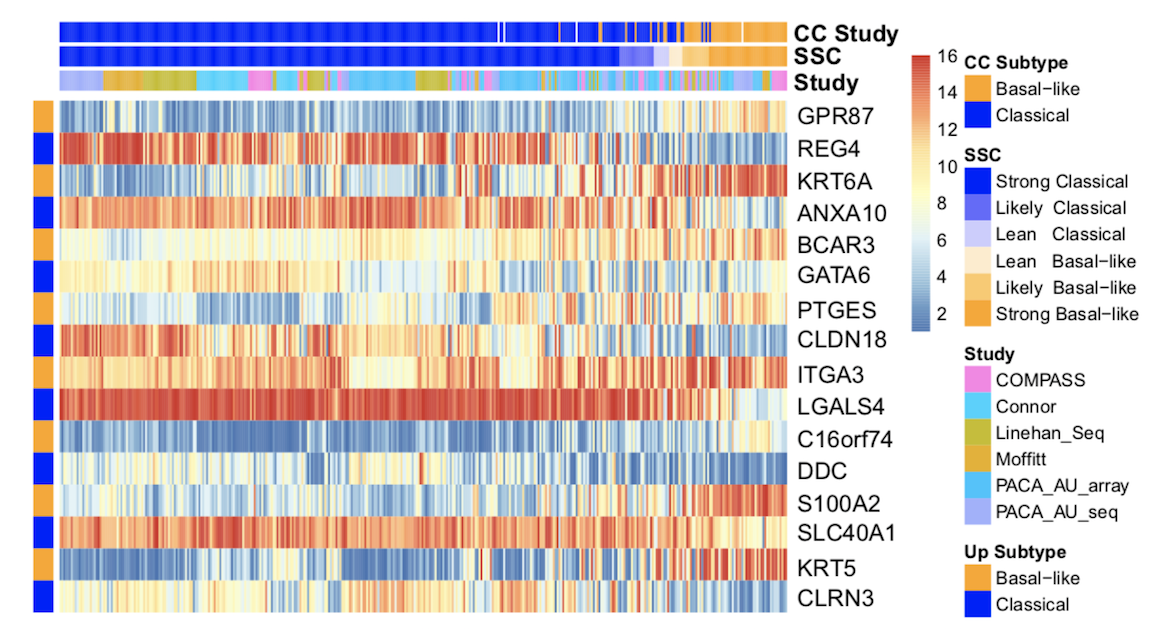
In 2015, the Yeh lab at UNC Lineberger proposed a novel two subtype gene expression-based molecular classification system, consisting of the “basal-like” and “classical” subtypes, faciliating “precision medicine” approaches in pancreatic cancer. Multiple subtyping systems for pancreatic cancer have been also proposed, and therefore consensus regarding subtypes for clinical decision-making has been elusive.
In a recent evaluation of existing subtyping systems in nine validation datasets, we found that the two-subtype system best explains survival outcomes and treatment response in a replicable manner across studies. We also show that basal-like patients are highly resistant to common first-line therapy FOLFIRINOX, suggesting alternative therapies for such patients up front.
Based on these results, as well as concepts from prior work, we have developed a high accuracy classifier called PurIST to predict pancreatic cancer subtype in new patients. This method is robust to normalization method, gene expression platform (NanoSting, RNA-seq), and sample collection process used (FFPE, FNA), and is highly accurate across a range of conditions. PurIST is currently undergoing CLIA certification for use in several upcoming clinical trials.
Github Repos
- PurIST: A clinically robust, single-sample classifier for tumor subtyping in pancreatic cancer. PurIST is for research purposes only and is NOT available for commercial use. To access PurIST please contact us with your github ID.
Related publications:
N.U. Rashid, X. L. Peng, C. Jin, R. A. Moffitt, K. E. Volmar, B. A. Belt, R. Z. Panni, T. M. Nywening, S. G. Herrera1, K. J. Moore1, S. G. Hennessey, A. B. Morrison, R. Kawalerski, A. Nayyar, A. E. Chang, B. Schmidt, H. J. Kim, D. C. Linehan, and J. J. Yeh. Purity independent subtyping of tumors (PurIST), a clinically robust, single-sample classifier for tumor subtyping in pancreatic cancer. Clinical Cancer Research, In Press, 2019
R. A. Moffitt, J. J. Yeh, and N.U. Rashid. Methods and compositions for prognostic and/or diagnostic subtyping of pancreatic cancer. US 20170233827A1, United States Patent and Trademark Office, 17 August 2017.
R. A. Moffitt, R. Marayati, E. L. Flate, K. E. Volmar, S. G. H. Loeza, K. A. Hoadley, N.U. Rashid, L. A. Williams, S. C. Eaton, A. H. Chung, et al. Virtual microdissection identifies distinct tumor-and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nature genetics, 47(10):1168, 2015
Developing statistical tools for high-throughput epigenomics
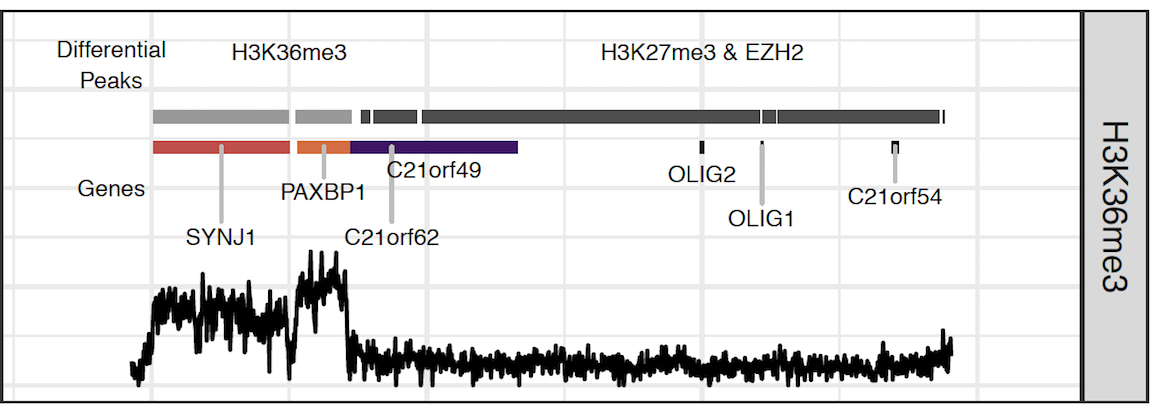
High throughput sequencing experiments such as ChIP-seq, ATAC-seq, and others are commonly utilized to characterize the human epigenome in fine resolution. A common goal in such experiments is to detect regions of the genome that are “enriched” for a particular type of epigenomic activity of interest, for example specific protein-DNA interactions such as transcription factor binding or histone modifications.
As the cost of sequencing has dropped, the experimental design of such experiments has become more complex and diverse, posing problems for methods optimized for specific data types or research questions. We develop efficient and robust methods to accurately detect regions of epigenomic activity, regardless of the type of experiment performed (short or broad enrichment), experimental design (multi-replicate and/or multi condition), or research question (peak calling, consensus peak calling, differential peak calling, or combinatiorial enrichment pattern detection).
We are in the early stages of developing deep learning models for single-cell epigenome experiments, such as single-cell ChIP-seq.
Github Repos
ZIMHMM: Consensus peak detection from multiple replicates
mixNBHMM: Flexible detection of broa and short differential regions of enrichment from multi-sample, multi-condition epigenomic experiments (ChIP-seq, ATAC-seq, DNase-seq, etc.). Detection of local combinational patterns of multiple epigenomic processes in the same sample or condition (chromatin state segmentation).
Related publications:
P. L. Baldoni, N.U. Rashid, and J. G. Ibrahim. Efficient detection and classification of epigenomic changes under multiple conditions. bioRxiv, 2019
P. L. Baldoni, N.U. Rashid, and J. G. Ibrahim. Improved detection of epigenomic marks with mixed effects hidden markov models. Biometrics, 2019
Y.-Y. Chiou, Y. Yang, N.U. Rashid, R. Ye, C. P. Selby, and A. Sancar. Mammalian period represses and de-represses transcription by displacing clock-bmal1 from promoters in a cryptochrome-dependent manner. Proceedings of the National Academy of Sciences, 113(41):E6072-E6079, 2016
N.U. Rashid, W. Sun, and J. G. Ibrahim. Some statistical strategies for dae-seq data analysis: variable selection and modeling dependencies among observations. Journal of the American Statistical Association, 109(505):78-94, 2014
B. Bernstein, E. Birney, I. Dunham, E. Green, C. Gunter, M. Snyder, et al. An integrated encyclopedia of dna elements in the human genome. Nature, 489(7414):57, 2012
N.U. Rashid, P. G. Giresi, J. G. Ibrahim, W. Sun, and J. D. Lieb. Zinba integrates local covariates with dna-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome biology, 12(7):R67, 2011
Multi-study replicability of prediction models
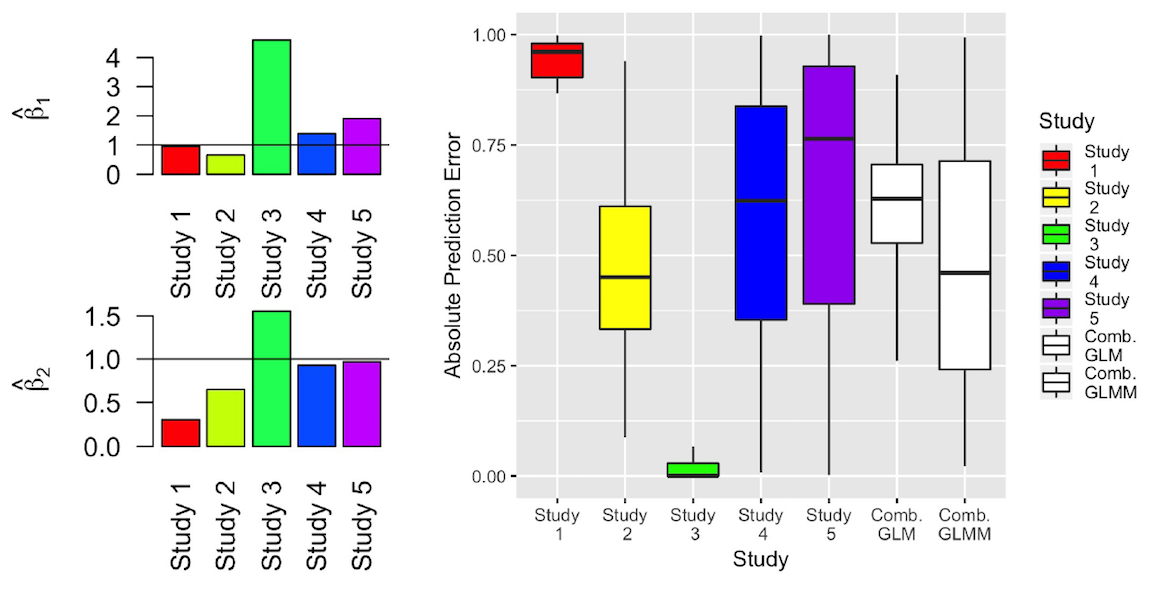
In the genomic era, the identification of gene signatures associated with disease is of significant interest. Such signatures are often used to predict clinical outcomes in new patients and aid clinical decision-making. However, recent studies have shown that gene signatures are often not replicable. This occurrence has practical implications regarding the generalizability and clinical applicability of such signatures.
To improve replicability, we develop novel approaches to select gene signatures from multiple datasets whose effects are consistently non-zero and account for between-study heterogeneity. We also build our models upon rank-based quantities (top scoring pairs), facilitating integration over different genomic datasets and prediction in new data. A formal R package is currently in development implementing these methods. We are currently extending this work to survival outcomes and also to tree-based machine learning models.
Related publications:
N.U. Rashid, X. L. Peng, C. Jin, R. A. Moffitt, K. E. Volmar, B. A. Belt, R. Z. Panni, T. M. Nywening, S. G. Herrera1, K. J. Moore1, S. G. Hennessey, A. B. Morrison, R. Kawalerski, A. Nayyar, A. E. Chang, B. Schmidt, H. J. Kim, D. C. Linehan, and J. J. Yeh. Purity independent subtyping of tumors (PurIST), a clinically robust, single-sample classifier for tumor subtyping in pancreatic cancer. Clinical Cancer Research, In Press, 2019
N.U. Rashid, Q. Li, J. J. Yeh, and J. G. Ibrahim. Modeling between-study heterogeneity for improved replicability in gene signature selection and clinical prediction. Journal of the American Statistical Association, In Press, 2019
Unsupervised clustering of RNA-seq data
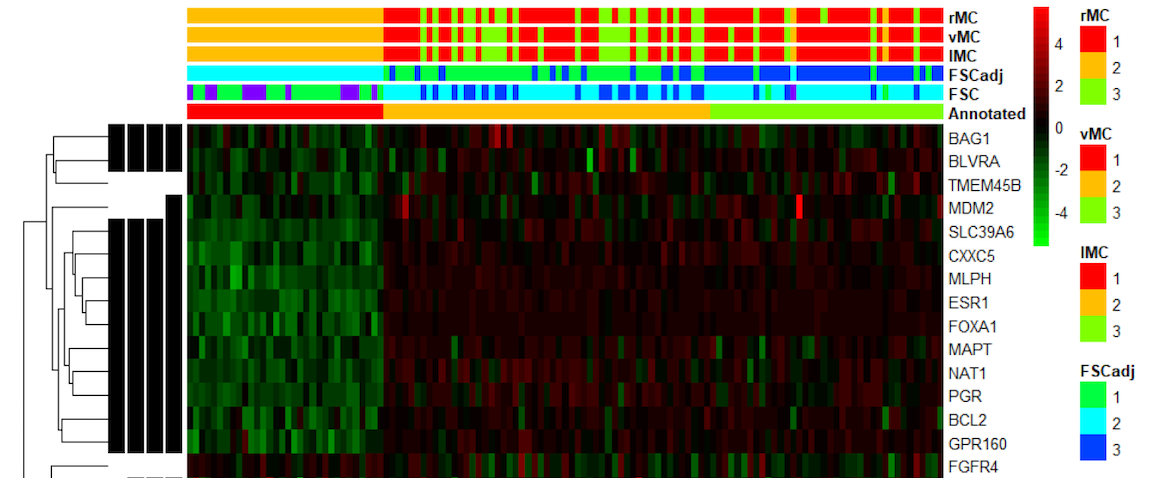
A common application of this in biomedical research is in delineating novel cancer subtypes from patient gene expression data, given a set of informative genes. However, it is typically unknown a priori what genes may be informative in discriminating between clusters, and what the optimal number of clusters is. In addition, few methods exist for unsupervised clustering of RNA-seq samples, and no method exists that can do so while simultaneously adjusting for between-sample global normalization factors, accounting for potential confounding variables (such as batch), and selecting cluster-discriminatory genes.
To address this issue, we develop novel a model-based clustering algorithm that utilizes a finite mixture of Negative Binomial regression models and employing a quadratic penalty method with a SCAD penalty. The maximization is done by penalized EM algorithm, allowing us to include normalization factors and confounders in our modeling framework. Given the fitted model, our framework allows for subtype prediction in new patients via posterior probabilities of cluster membership. Future directions of this work concern the extension to Deep Learning variational autoencoders, where we also focus on missing data problems in this setting.
Related talks:
- D. Lim, N.U. Rashid, and J.G.Ibrahim. FSCseq: Simultaneous Feature Selection and Clustering of RNA-Seq Data. JSM, 2019.
